Ribosome Competition
Reduce competition for ribosomes in RNA translation by splitting pools (orthogonal ribosomes)
When introducing a new (synthetic/circuit) gene into a cell, the new gene competes for resources with all of the existing (host) genes in the cell. Some of these resources include ribosome, DNA polymerase, RNA polymerase, and amino acids. In some extreme cases, the synthetic genes could hog the resources and not leave enough for the host genes, thus shutting down cell function and killing the cell. One method to try and reduce this competition for ribosomes is to create “orthogonal ribosomes,” which are ribosomes that only translate the circuit gene (and the circuit gene does not attract host ribosomes). This project uses the Ribosomal Flow Model (RFM) to form a dynamical model for orthogonal ribosome competition.
The RFM is a nonlinear tridiagonal dynamical system used in queuing theory that can represent a single strand of mRNA. Each cite (codon) has a probability of a ribosome being present (value x of the state). The flux from one codon to the next in the sequence is proportional to the current site’s occupancy times the next site’s vacancy.
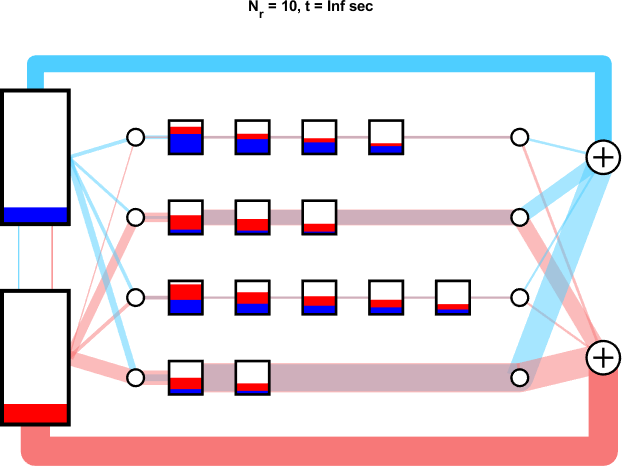
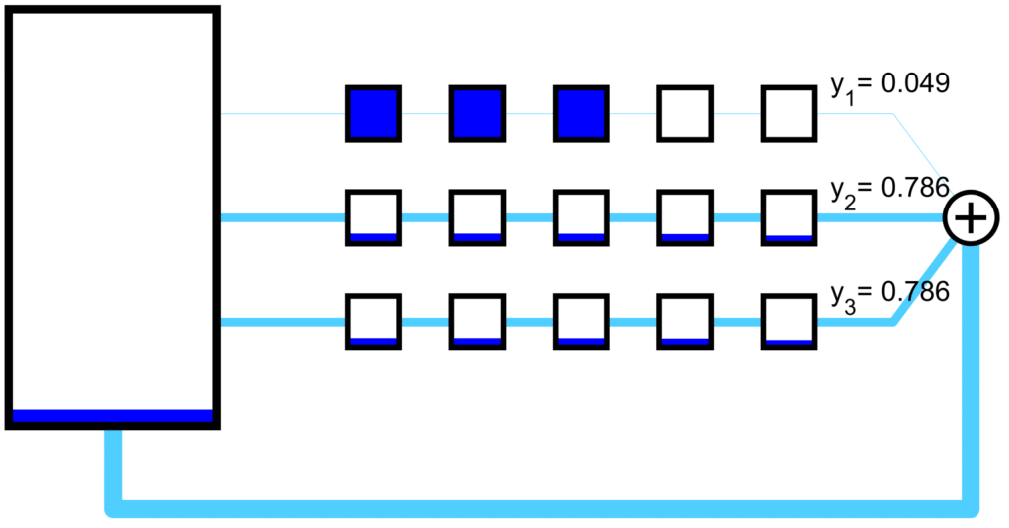
A set of mRNA strands (genes) may be connected to a common pool of ribosomes.
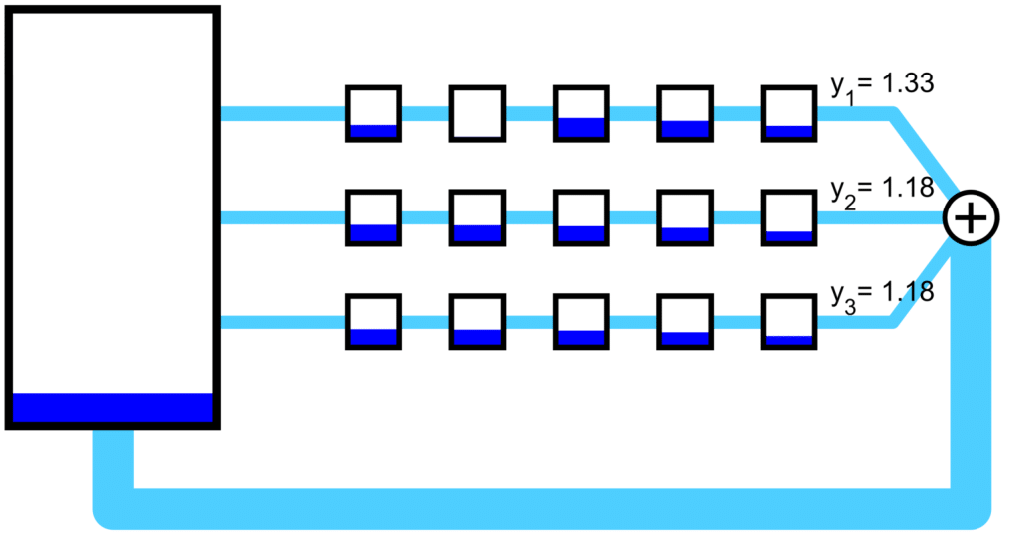
Orthogonal ribosomes may be introduced to translate the circuit genes. The host ribosomes (with host 16s rRNA) and circuit ribosomes (with mutated 16s rRNA) may each dissociate into and be created from ‘neutral’ribosomes by mass action, which abstract out the protein backbone of the ribosome. The Orthogonal Ribosomal Flow Model (ORFM) is stable and has a unique solution when codon occupancies are in [0, 1]. In the following figures, the host ribosomes are in blue, the circuit ribosomes are in red, and the ‘neutral’ ribosomes are in purple.
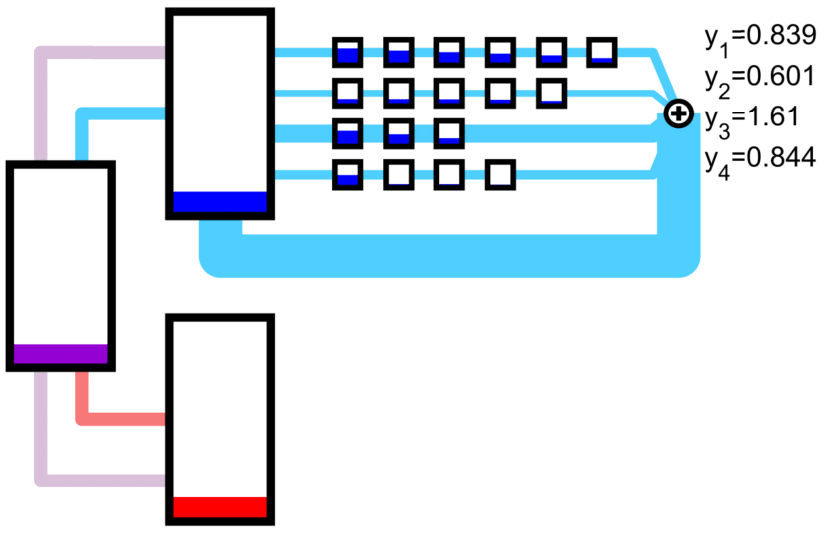
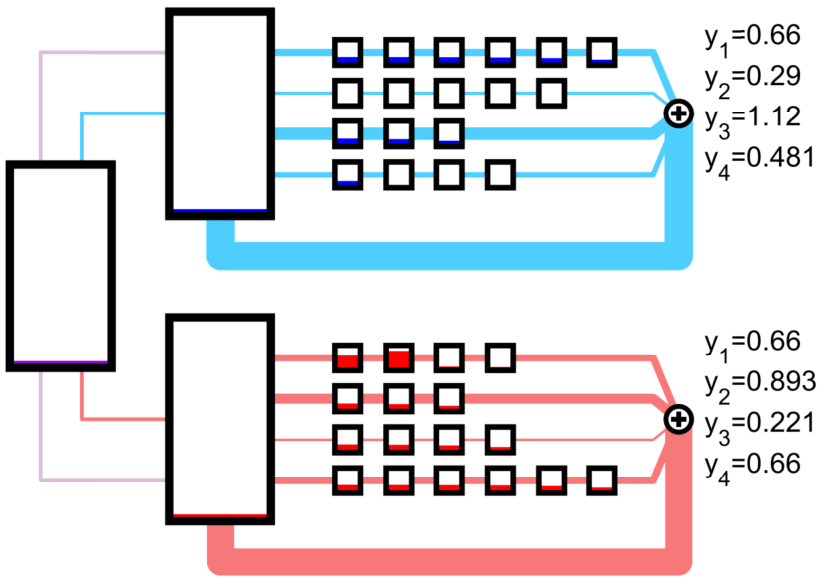
An nonconex optimization problem can be constructed to maximize a linear functional of the protein output rates by adjusting the creation rate of host and circuit ribosomes from the neutral ribosomes.
We also consider a self-inhibiting feedback controller that ensures orthogonal ribosomes are only created if they are needed. The golden circuit mRNA strand produces a protein which inhibits the formation of circuit ribosomes.


The ORFM model assumes a perfect rejection: host ribosomes will never translate circuit genes (and vice versa). The cross-talk RFM model allows for a varying-but-nonzero initiation rate between ribosomal species. Empirically the crosstalk ORFM model appears stable, but proving its stability remains an open question.